DeepMind introduced AlphaFold 3, the most recent iteration of its protein folding challenge.
AlphaFold 3, like its predecessors, primarily predicts how proteins fold primarily based on their amino acid sequences.
Proteins, the constructing blocks of all natural life, comprise lengthy chains of amino acids that fold like ‘origami’ into 3D constructions that decide their features.
AlphaFold makes use of machine studying to simulate the probably 3D construction a protein will undertake by folding.
Understanding how these constructions fold opens the door to deciphering the molecular mechanisms that underpin well being and illness.
Furthermore, proteins can turn out to be misfolded, a course of that not solely disrupts their regular perform but in addition contributes to the event of ailments akin to Alzheimer’s and Parkinson’s.
Misfolding can intervene with mobile well being by accumulating dysfunctional proteins that may harm cells and tissues.
The “protein folding drawback” that AlphaFold is attempting to crack refers to our understanding of how proteins are configured by amino acids.
Previous to machine studying strategies, the variety of potential configurations a protein can take is astronomically excessive, making it computationally intensive to foretell the proper construction by guide calculations or brute power alone.
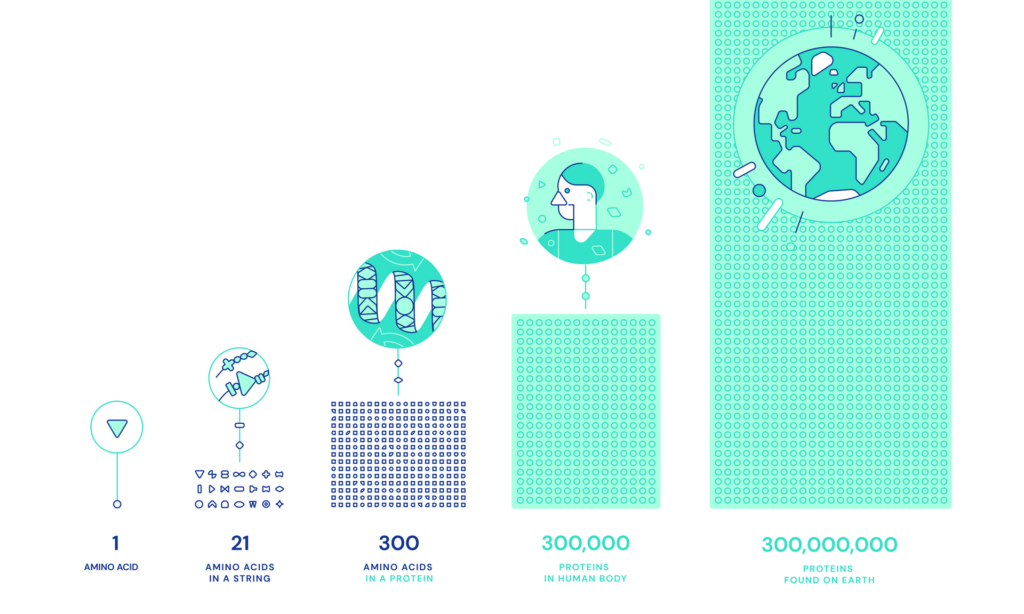
AlphaFold solves the problem of scale in predicting protein constructions utilizing deep studying.
The system makes use of neural networks educated on a database of identified protein constructions to deduce the 3D form of proteins from their amino acid sequences.
Introducing AlphaFold 3
DeepMind just lately introduced AlphaFold 3, which options an improved model of the Evoformer module, a part of the deep studying structure underpinning AlphaFold 2.
As soon as the Evoformer module processes enter molecules, AlphaFold 3 makes use of a novel diffusion community to assemble the anticipated constructions.
This community is just like these utilized in AI picture mills like DALL-E. It begins with a ‘cloud’ of atoms and iteratively refines the construction over a sequence of steps till it converges on a last, probably correct molecular configuration.
The AlphaFold 3 mannequin has advanced past proteins alone – it additionally incorporates data on DNA, RNA, and small molecules and might seize a few of their advanced interactions.
Isomorphic Labs, who collaborated with DeepMind on the AlphaFold 3 challenge, is already working with pharmaceutical firms, making use of the mannequin to real-world drug design challenges.
DeepMind has additionally launched the AlphaFold Server, a free and user-friendly platform that permits researchers to harness the ability of AlphaFold 3 with out intensive computational sources or experience in machine studying.
A brief historical past of the AlphaFold challenge
The AlphaFold challenge began in 2016 and led to 2018, shortly after AlphaGo’s historic victory in opposition to Lee Sedol, a prime worldwide Go participant.
In 2018, DeepMind debuted AlphaFold 1, the primary model of the AI system, on the CASP13 (Vital Evaluation of Protein Construction Prediction) problem.
This biennial competitors brings collectively analysis teams from around the globe to check the accuracy of their protein construction predictions in opposition to actual experimental knowledge.
AlphaFold 1 positioned first within the competitors, a large milestone in computational biology.
Two years later, at CASP14 in 2020, DeepMind introduced AlphaFold 2, demonstrating an accuracy so excessive that the scientific group thought of the protein-folding drawback basically solved.
AlphaFold 2’s efficiency was outstanding. It achieved a median accuracy rating of 92.4 GDT (World Distance Check) throughout all targets.
To place this into perspective, a rating of 90 GDT is taken into account aggressive with outcomes obtained from experimental strategies. The AlphaFold 2 strategies paper has since acquired over 20,000 citations, putting it among the many prime 500 most-cited papers throughout all scientific fields.
AlphaFold has been instrumental in quite a few novel analysis tasks, akin to finding out proteins which may degrade environmental pollution, akin to plastics, and enhancing our understanding of unusual tropical ailments like Leishmaniasis and Chagas.
In July 2021, DeepMind, in partnership with EMBL’s European Bioinformatics Institute (EMBL-EBI), launched the AlphaFold Protein Construction Database, which supplies entry to over 350,000 protein construction predictions, together with the whole human proteome.
This database has since been expanded to incorporate over 200 million constructions, protecting practically all cataloged proteins identified to science.
To this point, the AlphaFold Protein Construction Database has been accessed by over a million customers in over 190 nations, enabling discoveries in fields starting from drugs to agriculture and past.
AlphaFold 3 marks one other iteration for this best-in-class protein discovery and evaluation system.


